 Nematodes are the most abundant animals on earth and found in every ecological niche. They parasitize nearly every plant, insect and animal, including humans. They cause many diseases in humans such as hookworm, pin worm and intestinal roundworm infection (Ascariasis). Nematodes infect approximately 1/6 of humans in the world. Even though nematode infections are effectively treated with drugs, they are developing resistance to current nematicides, much like antibiotic resistant bacteria. To treat human intestinal nematode infections in the future, we need to develop novel drugs.
Nematodes are the most abundant animals on earth and found in every ecological niche. They parasitize nearly every plant, insect and animal, including humans. They cause many diseases in humans such as hookworm, pin worm and intestinal roundworm infection (Ascariasis). Nematodes infect approximately 1/6 of humans in the world. Even though nematode infections are effectively treated with drugs, they are developing resistance to current nematicides, much like antibiotic resistant bacteria. To treat human intestinal nematode infections in the future, we need to develop novel drugs.
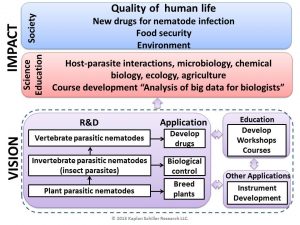
Understanding how nematodes infect has many important applications: 1- When nematodes infect insect, they can be used as biological control organisms for controlling agronomically important insect pests. 2- Plant parasitic nematodes are one of the major contributors of crop loss. They reduce yield, biomass, and quality of agronomically important crops. Nematodes were effectively controlled by the soil fumigant methyl bromide, which is banned. Currently, we need to develop environmentally friendly methods to control plant parasitic nematodes. 3- In order to control both plant and human parasitic nematodes, we need to understand how parasitic nematodes interact with their hosts. Nematode quorum sensing (QS) signals which regulate entry into a hardened larval stage are also important for nematode communication. In bacteria, QS signals reprogram animal and plant gene expression and immune system. Currently, we do not know whether nematode QS signals also reprogram host gene expression and immune systems. Understanding how nematode quorum sensing signals affect host parasite interaction provides us a platform to develop drugs that combat human parasitic nematodes, to breed nematode resistant plants and to control insect pests.
Dr. Kaplan’s next 5 year vision statement for nematode pheromone field was selected as prize winning essay by Genetics Society of America and published in “The GSA reporter” Winter 2014 issue.
Fatma Kaplan, Ph.D.

